pi = 3.1416
sqrt(2) = 1.4142
e = 2.7183ATOC 4815/5815
Linking Python Scripts Together - Week 5.5
CU Boulder ATOC
2026-01-01
Linking Python Scripts Together
Today’s Objectives
- Understand why we split code across multiple
.pyfiles - Import functions and classes from your own scripts
- Master the
if __name__ == "__main__"guard - Organize a small project into reusable modules
Reminders
Midterm Grades Posted!
- Check Canvas for scores and feedback
- Office hours this week for questions
Office Hours:
Will: Tu 11:15-12:15p Th 9-10a Aerospace Cafe
Aiden: M / W 330-430p DUAN D319
ATOC 4815/5815 Playlist

ATOC 4815/5815 Playlist
Spotify Playlist: ATOC4815
- This Lecture:
Hot Chip - Ready for the Fall / I Feel Better
- here → playlist

Why Multiple Files?
The One-Giant-Notebook Problem
You’ve been writing everything in one notebook or one script. That works at first…
But then a .py for our Lorenze system looks like this:
lorenz63_everything.py (400 lines)
├── import statements
├── Lorenz63 class
├── plotting functions
├── utility helpers
├── analysis code
└── if __name__ == "__main__": ... (maybe)Problems:
- Hard to find anything — scroll, scroll, scroll…
- Can’t reuse your
Lorenz63class in a new analysis without copy-pasting - One typo can break everything
- Collaborators can’t work on different parts at the same time
The Solution: Split Into Modules
A module is just a .py file that contains functions, classes, or variables.
my_project/
├── lorenz63.py ← the Lorenz63 class
├── plotting.py ← plotting helper functions
├── utils.py ← small utility functions
└── run_experiment.py ← the main script that ties it all togetherBenefits:
- Each file has one job (single responsibility)
- Reuse
lorenz63.pyin any future project - Easier to read, test, and debug
- Multiple people can work on different files
You already do this! Every time you write import numpy, you’re importing a module someone else wrote.
Importing Your Own Code
Review: Importing Built-In Modules
You’ve been doing this since week 1:
The same syntax works for your own .py files!
Importing From Your Own File
Step 1: Write a file called conversions.py:
Three Import Styles
All three work. Each has trade-offs:
Import the module:
Verbose but clear where things come from.
Import specific names:
Shorter. You see exactly what you use.
General Rule: Use from module import name1, name2 for most cases. Use import module when you use many things from it (like numpy).
Common Error: ModuleNotFoundError
Predict the output:
ModuleNotFoundError: No module named 'conversions'Why? Python can’t find conversions.py. Most common causes:
| Cause | Fix |
|---|---|
| File doesn’t exist | Create conversions.py |
| File is in a different folder | Move it, or adjust sys.path |
| Typo in the name | import conveRsions vs import conversions |
| Running from wrong directory | cd to the project folder first |
Check Your Understanding
Given this project layout:
weather_project/
├── conversions.py ← has celsius_to_fahrenheit()
├── analysis.py ← your main script
└── data/
└── temps.csvWhich import works in analysis.py?
Answer: Option A
- A works because
conversions.pyis in the same folder asanalysis.py - B would work if you’re running from outside
weather_project/and it’s a package - C is wrong — you import modules (files), not functions directly
The __name__ Guard
The Problem: Code That Runs on Import
Imagine conversions.py has some test code at the bottom:
# conversions.py
def celsius_to_fahrenheit(temp_c):
return temp_c * 9/5 + 32
def fahrenheit_to_celsius(temp_f):
return (temp_f - 32) * 5/9
# Quick test
print("Testing conversions...")
print(f"0°C = {celsius_to_fahrenheit(0)}°F")
print(f"100°C = {celsius_to_fahrenheit(100)}°F")
print("All tests passed!")Why Does This Happen?
Key insight: When Python imports a .py file, it executes the entire file from top to bottom.
- Function/class definitions → stored for later use (good!)
- Any code at the top level → runs immediately (often bad!)
import conversions
│
├─ def celsius_to_fahrenheit(...) ← defined, stored ✓
├─ def fahrenheit_to_celsius(...) ← defined, stored ✓
├─ print("Testing conversions...") ← RUNS immediately ✗
├─ print(f"0°C = ...") ← RUNS immediately ✗
└─ print("All tests passed!") ← RUNS immediately ✗This is why we need the __name__ guard.
The __name__ Variable
Every Python file has a built-in variable called __name__.
It has two possible values:
| Situation | __name__ equals |
|---|---|
You run the file directly: python my_file.py |
"__main__" |
Someone imports the file: import my_file |
"my_file" |
Demo:
This is how a file knows whether it’s being run or imported!
The Guard Pattern
Wrap any “run only when executed directly” code in this block:
# conversions.py
def celsius_to_fahrenheit(temp_c):
return temp_c * 9/5 + 32
def fahrenheit_to_celsius(temp_f):
return (temp_f - 32) * 5/9
if __name__ == "__main__":
# This only runs when you do: python conversions.py
print("Testing conversions...")
print(f"0°C = {celsius_to_fahrenheit(0)}°F")
print(f"100°C = {celsius_to_fahrenheit(100)}°F")
print("All tests passed!")Now:
| Action | What happens |
|---|---|
python conversions.py |
Functions defined + tests run |
from conversions import celsius_to_fahrenheit |
Functions defined, tests skipped |
Common Error: Forgetting the Guard
Predict what happens:
# euler.py
import numpy as np
class Lorenz63:
def __init__(self, sigma=10, rho=28, beta=8/3, dt=0.01):
self.sigma, self.rho, self.beta, self.dt = sigma, rho, beta, dt
def tendency(self, state):
x, y, z = state
return np.array([self.sigma*(y-x), self.rho*x - y - x*z, x*y - self.beta*z])
def step(self, state):
return state + self.tendency(state) * self.dt
# "Quick test" without guard
model = Lorenz63()
state = np.array([1.0, 1.0, 1.0])
for _ in range(1000):
state = model.step(state)
print(f"Final state: {state}")Check Your Understanding
Which version is correct?
Version A:
Answer: Version B
- Version A:
from stats import anomalywould print output every time - Version B:
from stats import anomalyjust gives you the function, cleanly - Version B still works as a standalone script:
python stats.pyruns the test
Building a Real Project
Project Structure: Weather Toolkit
Let’s build a small multi-file project step by step:
weather_toolkit/
├── conversions.py ← temperature unit conversions
├── stats.py ← statistical analysis functions
├── plotting.py ← visualization helpers
└── run_analysis.py ← main script that ties it all togetherDesign principle: Each file has one job.
| File | Responsibility |
|---|---|
conversions.py |
Unit conversions (C↔︎F↔︎K) |
stats.py |
Anomalies, climatology, running mean |
plotting.py |
Standard plot templates for the course |
run_analysis.py |
Load data, call functions, produce output |
This is how real scientific code is organized — the same pattern scales from homework to research.
Step 1: conversions.py
# conversions.py
"""Temperature unit conversion utilities."""
def celsius_to_fahrenheit(temp_c):
"""Convert Celsius to Fahrenheit."""
return temp_c * 9/5 + 32
def celsius_to_kelvin(temp_c):
"""Convert Celsius to Kelvin."""
return temp_c + 273.15
def wind_speed_knots_to_ms(knots):
"""Convert wind speed from knots to m/s."""
return knots * 0.514444
if __name__ == "__main__":
# Quick sanity checks
assert celsius_to_fahrenheit(0) == 32
assert celsius_to_fahrenheit(100) == 212
assert celsius_to_kelvin(0) == 273.15
print("conversions.py: all checks passed")Notice:
- Docstrings on every function (good practice!)
if __name__guard withassertstatements for self-testing- Running
python conversions.pyverifies the module works
Step 2: stats.py
# stats.py
"""Statistical analysis functions for atmospheric data."""
import numpy as np
def compute_anomaly(data, axis=None):
"""Subtract the mean to get anomalies."""
climatology = data.mean(axis=axis, keepdims=True)
return data - climatology
def running_mean(data, window):
"""Compute running mean with given window size."""
kernel = np.ones(window) / window
return np.convolve(data, kernel, mode='valid')
def find_extremes(data):
"""Return dict with min, max, and their indices."""
return {
'min_val': data.min(),
'max_val': data.max(),
'min_idx': data.argmin(),
'max_idx': data.argmax(),
}
if __name__ == "__main__":
test_data = np.array([1.0, 2.0, 3.0, 4.0, 5.0])
anom = compute_anomaly(test_data)
assert np.isclose(anom.mean(), 0.0)
print("stats.py: all checks passed")Step 3: plotting.py
# plotting.py
"""Standard plotting templates for ATOC 4815."""
import matplotlib.pyplot as plt
def plot_timeseries(time, data, ylabel, title, label=None, ax=None):
"""Create a labeled time series plot."""
if ax is None:
fig, ax = plt.subplots(figsize=(9, 4))
ax.plot(time, data, linewidth=2, label=label)
ax.set_xlabel('Time')
ax.set_ylabel(ylabel)
ax.set_title(title)
ax.grid(True, alpha=0.3)
if label:
ax.legend()
return ax
def plot_comparison(time, datasets, labels, ylabel, title):
"""Plot multiple time series on the same axes."""
fig, ax = plt.subplots(figsize=(9, 4))
for data, label in zip(datasets, labels):
ax.plot(time, data, linewidth=2, label=label)
ax.set_xlabel('Time')
ax.set_ylabel(ylabel)
ax.set_title(title)
ax.legend()
ax.grid(True, alpha=0.3)
plt.tight_layout()
return fig, ax
if __name__ == "__main__":
import numpy as np
t = np.linspace(0, 24, 100)
y = 15 + 8 * np.sin(t * np.pi / 12)
plot_timeseries(t, y, 'Temp (°C)', 'Test Plot')
plt.show()
print("plotting.py: visual check passed")Step 4: run_analysis.py
# run_analysis.py
"""Main analysis script — ties all modules together."""
import numpy as np
from conversions import celsius_to_fahrenheit
from stats import compute_anomaly, running_mean, find_extremes
from plotting import plot_timeseries, plot_comparison
# --- Generate synthetic data ---
np.random.seed(42)
hours = np.arange(0, 72) # 3 days
temp_c = 15 + 8 * np.sin((hours - 6) * np.pi / 12) + np.random.randn(72) * 2
# --- Analysis ---
temp_f = celsius_to_fahrenheit(temp_c)
anomalies = compute_anomaly(temp_c)
smooth = running_mean(temp_c, window=6)
extremes = find_extremes(temp_c)
print(f"Max temp: {extremes['max_val']:.1f}°C at hour {extremes['max_idx']}")
print(f"Min temp: {extremes['min_val']:.1f}°C at hour {extremes['min_idx']}")
# --- Visualization ---
plot_comparison(
hours,
[temp_c, np.pad(smooth, (2, 3), constant_values=np.nan)],
['Raw', '6-hr Running Mean'],
'Temperature (°C)',
'72-Hour Boulder Temperature Analysis'
)
import matplotlib.pyplot as plt
plt.savefig('boulder_analysis.png', dpi=150, bbox_inches='tight')
plt.show()
print("Analysis complete!")Look how clean this is! The main script reads almost like English because each function has a clear name and lives in a focused module.
The Import Flow
When you run python run_analysis.py, here’s what happens:
python run_analysis.py
│
├─ import numpy as np ← from installed packages
├─ from conversions import celsius_to_fahrenheit
│ └─ Python executes conversions.py
│ ├─ defines celsius_to_fahrenheit ← kept ✓
│ ├─ defines celsius_to_kelvin ← kept (not imported, but defined)
│ └─ if __name__ == "__main__": ... ← SKIPPED (name is "conversions")
├─ from stats import compute_anomaly, ...
│ └─ Python executes stats.py
│ ├─ import numpy as np ← numpy loaded (cached)
│ ├─ defines compute_anomaly ← kept ✓
│ └─ if __name__ == "__main__": ... ← SKIPPED
├─ from plotting import ...
│ └─ ...same pattern...
│
└─ Your analysis code runsEvery imported file is executed once, top to bottom. The __name__ guard prevents side effects.
Import Pitfalls
Circular Imports
What happens here?
ImportError: cannot import name 'get_temp' from partially initialized
module 'temperature' (most likely due to a circular import)What went wrong?
- Python starts loading
wind.py - Line 1:
from temperature import get_temp→ Python pauseswind.pyand starts loadingtemperature.py - Line 1 of
temperature.py:from wind import wind_chill→ butwind.pyisn’t done yet! wind_chilldoesn’t exist yet → crash
Fixing Circular Imports
The rule: import dependencies must flow one way — like a river, not a loop.
Before (broken): each file imports from the other
wind.py ──imports──▶ temperature.py
▲ │
└────────imports─────────┘ ← CYCLE!After (fixed): extract the shared piece into a third file
core.py ◀── wind.py ◀── temperature.py ← one direction, no cycle ✓Import Order Convention
PEP 8 (the Python style guide) recommends this order:
Why?
- Separates “always available” from “need to install” from “our code”
- Makes it easy to see dependencies at a glance
- Most linters (like
flake8) will warn if you mix them up
Simple rule: stdlib → third-party → yours, with a blank line between each group.
Common Error: Shadowing Module Names
Predict the output:
AttributeError: module 'math' has no attribute 'sqrt'Why? Python found your math.py before the built-in math module!
Dangerous file names to avoid:
| Don’t name your file | It shadows |
|---|---|
math.py |
import math |
random.py |
import random |
numpy.py |
import numpy |
test.py |
import test (built-in) |
statistics.py |
import statistics |
Fix: Rename your file to something specific: my_math_utils.py, weather_stats.py, etc.
The __init__.py File
When your Python project grows beyond loose .py files into an organized folder, you’ll see a special file called __init__.py. What does it do, and when do you actually need it?
“Wait, I can already import files without this…” — Yes! If you’re inside a folder, Python finds sibling files automatically:
But when you use dot notation to reach into a folder from outside, Python needs __init__.py:
lorenz_project/
├── __init__.py ← without this: ModuleNotFoundError
├── integrators.py
├── lorenz63.py
└── plotting.pyWithout __init__.py:
With __init__.py (even empty):
Rule: __init__.py is needed when you reference a folder with dot notation — i.e., when you treat it as a package rather than just a directory you happen to be sitting in.
__init__.py — More Than Just Empty
An empty __init__.py is fine to start, but it can also do useful things:
1. Convenience imports — let users skip the submodule path:
Now instead of the long path:
users can just write:
2. Package-level metadata:
3. Control what from package import * exports:
For this lab: an empty __init__.py is all you need. But as your projects grow, this file becomes your package’s front door.
Lab: Lorenz63 Ensemble Experiment
Predictability Depends on Location
The Lorenz attractor has regions where forecasts stay accurate and regions where they immediately diverge.
Your goal: Build a multi-file project that produces this figure:
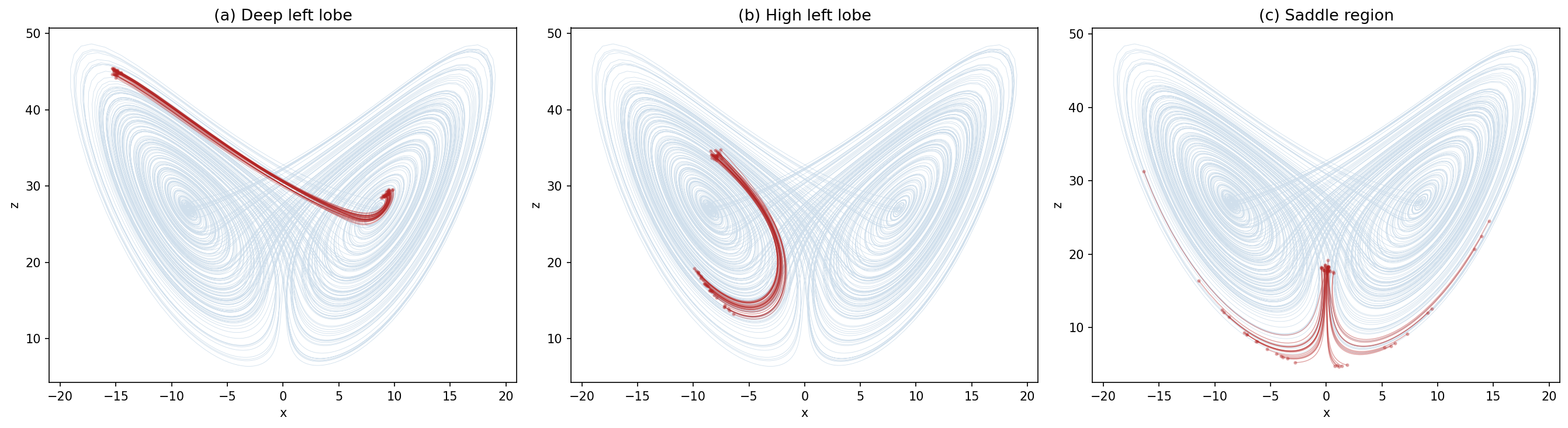
Three panels showing ensembles of trajectories started from different regions of the attractor:
| Panel | Starting region | What you’ll see |
|---|---|---|
| (a) Deep left lobe [-15,1,45] | Deep in the left wing | Ring of uncertainty barely grows — highly predictable |
| (b) High left lobe [-8,-3,34] | Upper part of left wing, near transition | ensemble slightly diverves - ** medium predictability ** |
| (c) Saddle region [0,0,18] | Between the two wings | Ensemble explodes — some go left, some go right — no predictability |
This is the key insight of chaos: not all regions of a system are equally predictable. Weather models face exactly this challenge.
Your Project Structure
Refactor your midterm code into this multi-file project:
lorenz_project/
├── __init__.py ← empty, makes this a package
├── integrators.py ← euler_step() and integrate()
├── lorenz63.py ← Lorenz63 class with run() and run_ensemble()
├── plotting.py ← plot_attractor(), plot_ensemble(), plot_ensemble_panels()
└── run_lorenz_ensemble.py ← driver script — produces the 3-panel figureWho imports whom? Read each arrow as “imports from”:
run_lorenz_ensemble.py
├── imports from ──▶ lorenz63.py
│ └── imports from ──▶ integrators.py
└── imports from ──▶ plotting.pyNotice: arrows only point downward/right — no file imports from a file that imports it back. That’s what “no circular imports” means.
Starter files are provided — each has docstrings, hints, and TODO markers. Fill them in.
File 1: integrators.py
What to implement:
| Function | Signature | What it does |
|---|---|---|
euler_step |
(state, tendency_fn, dt) → state |
One Forward Euler step |
integrate |
(state0, tendency_fn, dt, n_steps) → trajectory |
Full time loop, returns shape (n_steps+1, n_vars) |
File 2: lorenz63.py
What to implement:
| Method | What it does |
|---|---|
__init__(sigma, rho, beta) |
Store parameters |
tendency(state) |
Return \([dx/dt,\; dy/dt,\; dz/dt]\) |
run(state0, dt, n_steps) |
Integrate one trajectory (calls integrate from integrators.py) |
run_ensemble(ics, dt, n_steps) |
Integrate many trajectories from an array of initial conditions |
The ensemble method — implement it two ways:
- Nested for loop: loop over members, call
self.run()for each one - Vectorized: single loop over time steps, advance all members at once
# Method 1: loop over members (straightforward)
for i in range(n_members):
ensemble[i] = self.run(ics[i], dt, n_steps)
# Method 2: loop over time only (fast!)
# states shape: (n_members, 3) — all members at once
for t in range(n_steps):
states = states + vectorized_tendency(states) * dtFile 3: plotting.py
What to implement:
| Function | What it does |
|---|---|
plot_attractor(ax, trajectory) |
Plot one trajectory in \(x\)-\(z\) phase space (the butterfly) |
plot_ensemble(ax, ensemble, reference) |
Plot reference attractor (light) + ensemble members (bold) |
plot_ensemble_panels(ensemble_list, reference, titles) |
Create the 3-panel figure, one panel per starting region |
Plotting tips:
- Use
trajectory[:, 0]for \(x\) andtrajectory[:, 2]for \(z\) - Reference attractor: light color, low alpha (
alpha=0.3) - Ensemble members: bold color like
"firebrick", higher alpha plt.savefig(path, dpi=150, bbox_inches='tight')to save
File 4: run_lorenz_ensemble.py
The driver script. This ties everything together:
- Create a
Lorenz63model with default parameters - Spin up a reference trajectory (2000 steps to get on the attractor, then 10000 more)
- Pick 3 starting points from the reference — one in each region:
- Deep left lobe: find a point where \(x < -10\) and \(z < 25\)
- High left lobe: find a point where \(x < -5\) and \(z > 35\)
- Saddle: find a point where \(|x| < 2\)
- For each starting point, create ~30 nearby initial conditions by adding small perturbations (
np.random.randn(30, 3) * 0.5) - Run
model.run_ensemble()for each cloud - Call
plot_ensemble_panels()to make the 3-panel figure - Save to
lorenz_ensemble_predictability.png
Deliverables Checklist
integrators.py— runpython -m lorenz_project.integratorsand confirm it prints"all checks passed!"(tests Euler on \(dy/dt = -y\), checks answer ≈ \(e^{-1}\))lorenz63.py— runpython -m lorenz_project.lorenz63and confirm it prints"all checks passed!"(checks trajectory shape is(1001, 3)and ensemble shape is(5, 1001, 3))plotting.py— runpython -m lorenz_project.plotting— a plot window should appear with a random-walk line. If you see a line, it works.run_lorenz_ensemble.py— runpython -m lorenz_project.run_lorenz_ensemble— produces and saveslorenz_ensemble_predictability.png- Every file has an
if __name__ == "__main__":guard with tests inside it run_ensembleimplemented with Method 1 (nested loop); Method 2 (vectorized) for bonus- The figure —
lorenz_ensemble_predictability.pngwith three panels showing how predictability varies across the attractor
Grading:
- Working multi-file structure with correct imports: 40%
- Ensemble method (Method 1): 30%
- Final 3-panel figure: 20%
- Vectorized ensemble (Method 2, bonus): 10%
What You’ll Discover
When your figure is done, you’ll see:
(a) Deep left lobe — The ensemble starts tight and stays together. Trajectories orbit the left wing in sync. The ring of uncertainty barely grows. Highly predictable.
(b) High left lobe — The ensemble starts tight but distorts. Some members continue looping left, others begin transitioning to the right lobe. The ring stretches into banana and boomerang shapes. Partially predictable — you know roughly where, but not which lobe.
(c) Saddle region — The ensemble starts tight but immediately explodes. Some trajectories go left, some go right. This is where the two wings meet and tiny differences get maximally amplified. No predictability.
This is exactly the problem weather forecasters face: some atmospheric states are inherently more predictable than others. Ensemble forecasting reveals where confidence is warranted and where it isn’t.
Summary
Key Takeaways
Split code into modules — each
.pyfile should have one clear jobImport styles:
import module→module.function()(explicit)from module import function→function()(convenient)from module import *→ avoid (namespace pollution)
Always use
if __name__ == "__main__":to guard code that should only run when the file is executed directlyAvoid pitfalls:
- Don’t name files after built-in modules
- Keep import dependencies one-directional (no circular imports)
- Follow PEP 8 import ordering: stdlib → third-party → local
This pattern scales — from homework to research code to production software
The __name__ Guard Cheat Sheet
# my_module.py
# --- Imports at the top ---
import numpy as np
# --- Functions and classes ---
def my_function():
...
class MyClass:
...
# --- Guard: only runs when executed directly ---
if __name__ == "__main__":
# Tests, demos, or standalone behavior
result = my_function()
print(f"Self-test: {result}")Memorize this template. Every .py file you write for this course (and beyond) should follow it.
Looking Ahead
Next lectures:
- Week 6: Git for scientists — version control your projects
- Week 7: Tabular data with Pandas
This week’s lab:
- Complete the
lorenz_project/files - Produce the 3-panel ensemble predictability figure
- Submit: all
.pyfiles + the saved figure
Pro tip: Build and test one file at a time. Run each self-test before moving to the next file.
Questions?
Key things to remember:
- A module is just a
.pyfile importexecutes the file,__name__controls what runs- One-way dependencies, no circles
- Build bottom-up:
integrators→lorenz63→plotting→run_lorenz_ensemble
You’ve got this! You already wrote all the hard code for the midterm. Now you’re organizing it and extending it.
Contact
Prof. Will Chapman
wchapman@colorado.edu
willychap.github.io
ATOC Building, CU Boulder
See you next week!

ATOC 4815/5815 - Week 5.5